As we’re wrapping up what may be the worst year in recent global memory, especially geopolitically, let’s take a moment to review some more positive things that came up at Lawnchair in 2016.
Headed home

Alternate subtitle: Go West
This was a quiet year on the blog, with only 18 posts compared with the roughly thirty per year in 2014-2015. The major reason for the silence was that I moved from Kazakhstan back to the US to join the Anthropology Department at Vassar College in New York. With all the movement there was less time to blog. Much of the second half of 2016 was spent setting up the Biological Anthropology Lab at Vassar, which will focus on “virtual” anthropology, including 3D surface scanning…

Cast of early Homo cranium KNM-ER 1470 and 3D surface scan made in the lab using an Artec Spider.
… and 3D printing.

A gibbon endocast, created from a CT scan using Avizo software and printed on a Zortrax M200.
This first semester stateside I reworked my ‘Intro to Bio Anthro’ and ‘Race’ courses, which I think went pretty well being presented to an American audience for the first time. The latter class examines human biological variation, situating empirical observations in modern and historical social contexts. This is an especially important class today as 2016 saw a rise in nationalist and racist movements across the globe. Just yesterday Sarah Zhang published an essay in The Atlantic titled, “Will the Alt-right peddle a new kind of racist genetics?” It’s a great read, and I’m pleased to say that in the Race class this semester, we addressed all of the various social and scientific issues that came up in that piece. Admittedly though, I’m dismayed that this scary question has to be raised at this point in time, but it’s important for scholars to address and publicize given our society’s tragically short and selective memory.
So the first semester went well, and next semester I’ll be teaching a seminar focused on Homo naledi and a mid-level course on the prehistory of Central Asia. The Homo naledi class will be lots of fun, as we’ll used 3D printouts of H. naledi and other hominin species to address questions in human evolution. The Central Asia class will be good prep for when I return to Kazakhstan next summer to continue the hunt for human fossils in the country.
Osteology is still everywhere
A recurring segment over the years has been “Osteology Everywhere,” in which I recount how something I’ve seen out and about reminds me of a certain bone or fossil. Five of the blog 18 posts this year were OAs, and four of these were fossiliferous: I saw …

Anatomy terminology hidden in 3D block letters,

Hominin canines in Kazakhstani baursaki cakes,

The Ardipithecus ramidus ilium in Almaty,

A Homo naledi juvenile femur head in nutmeg,

And a Homo erectus cranium on a Bangkok sidewalk. As I’m teaching a fossil-focused seminar next semester, OA will probably become increasingly about fossils, and I’ll probably get my students involved in the fun as well.
New discoveries and enduring questions
The most-read post on the blog this year was about the recovery of the oldest human Nuclear DNA, from the 450,000 year old Sima de los Huesos fossils. My 2013 prediction that nuclear DNA would conflict with mtDNA by showing these hominins to be closer to Neandertals than Denisovans was shown to be correct.

These results are significant in part because they demonstrate one way that new insights can be gained from fossils that have been known for years. But more intriguingly, the ability of researchers to extract DNA from exceedingly old fossils suggests that this is only the tip of the iceberg.
The other major discoveries I covered this year were the capuchin monkeys who made stone tools and the possibility that living humans and extinct Neandertals share a common pattern of brain development.

An unrelated image from 2016 that makes me laugh.
The comparison between monkey-made and anthropogenic stone tools drives home the now dated fact that humans aren’t the only rock-modifiers. But the significance for the evolution of human tool use is less clear cut – what are the parallels (if any) in the motivation and modification of rocks between hominins and capuchins, who haven’t shared a common ancestor for tens of millions of years? I’m sure we’ll hear more on that in the coming years.
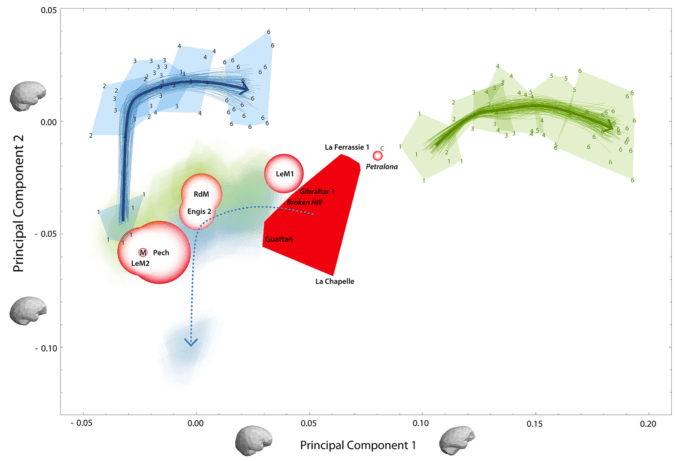
In the case of whether Neandertal brain development is like that of humans, I pointed out that new study’s results differ from previous research probably because of differences samples and methods. The only way to reconcile this issue is for the two teams of researchers, one based in Zurich and the other in Leipzig, to come together or for a third party to try their hand at the analysis. Maybe we’ll see this in 2017, maybe not.
There were other cool things in 2016 that I just didn’t get around to writing about, such as the publication of new Laetoli footprints with accompanying free 3D scans, new papers on Homo naledi that are in press in the Journal of Human Evolution, and new analysis of old Lucy (Australopithecus afarensis) fossils suggesting that she spent a lifetime climbing trees but may have sucked at it. But here’s hoping that 2017 tops 2016, on the blog, in the fossil record, and basically on Earth in general.