What makes the human brain special, and how did it change throughout our evolutionary history? One way to answer this question by comparing actual brains or MRI scans of living animals. But only fossils can show what changed and when over the past several million years, and sadly brains are basically an elaborately congealed soup that doesn’t stay fresh upon death, so they never fossilize. Happily, though, bones can preserve for millions of years, and they are literally molded by their soft and squishy surroundings. As the brain grows, it pushes outward against the inner surface of the skull, which can save the scars of the submerged cerebrum: nerds like me call these impressions an “endocast.”

Nicole Labra and Antoine Balzeau have led a cool study, hot off the press, examining what such endocasts can tell us about the underlying brain anatomy. Importantly, they show how difficult it is to clearly and consistently identify many brainy boundaries. This is very salient in ‘paleoneurology,’ the study of brain evolution especially based off endocasts: the problem probably best illustrated by the nearly century-long debate about the natural endcoast of the “Taung child” fossil (Australopithecus africanus).
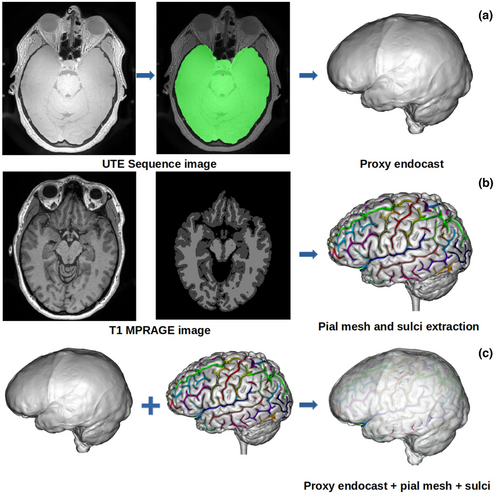
Labra & colleagues used a clever approach to address this paleontological and epistemological problem. They first generated an endocast directly associated with its brain from an MRI scan of a living human, allowing them see precisely where specific brain folds (“sulci”) lay relative to the endocast surface. They then asked a bunch of researchers—myself included—to try to identify sulci on the endocast, and then looked at how our responses compared to both one another’s and to the actual, known sulcus positions.
Their analysis showed that we varied quite a bit in our identifications on the endocast. As Emiliano Bruner (who also participated) discusses in his blog post, we tended to identify the stronger impressions toward the bottom and sides of the endocast better and more consistently. Some of this variability and uncertainty among researchers is due to the faintness and incompleteness of many brain impressions, and some due to biased expectations about where a given sulcus “should” be based on our previous experiences and published references.
When Antoine Balzeau first contacted me about this project, I was just beginning to dabble in “paleoneurology,” learning some brain anatomy for the first time for a description of an old Australopithecus endocast called “MLD 3.” I initially thought MLD 3 would be a quick and simple study—boy was I spectacularly disappointed!
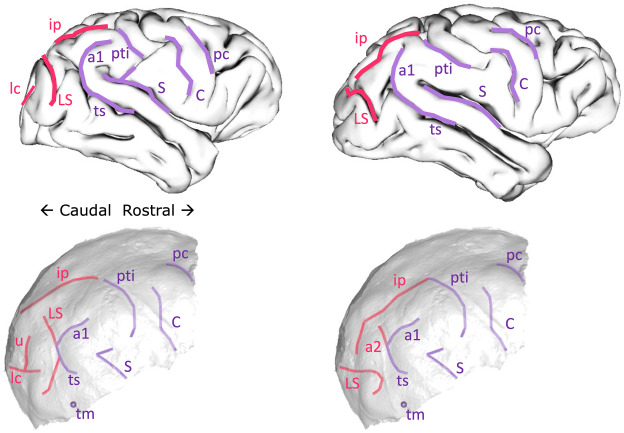
Probably reflecting observer bias and desire for definitive results, we initially interpreted the endocast impressions on MLD 3 as representing a ‘human-like’ anatomy that is super rare in living chimpanzees (namely the “LS” depicted in the right half of the figure above). The researchers who peer-reviewed the first draft of our paper, though, suggested we to be more cautious in our interpretations; one reviewer outright disagreed with us in support of a more ‘ape-like’ interpretation (left half of the figure above). The review process alone underscored the subjectivity and uncertainty in analyzing endocasts. In the end we presented both interpretations, and I honestly don’t know which (if either) is most likely to be correct. So the study by Labra and colleagues provides a nice empirical illustration of this cranial conundrum.
Fortunately, researchers are developing methods to help identify brain structures on endocasts. Jean Dumoncel, Edwin de Jager, and Amélie Beaudet among others have done some really impressive work looking at variability in both brains (for instance here) and endocasts (for instance here). By using computer-based 3D data and methods, these researchers have shown where many brain sulci tend to be located (see here). By developing a better understanding of variation in where sulci sit on an endocast, we can have a better idea of which sulci might be represented on fossil endocasts, which in turn can tell us about the brains of our extinct relatives. Edwin and Amélie presented a very cool new analysis of Australopithecus/Paranthropus boisei endocasts, building off this digital approach, at the recent ESHE conference. And as noted in our MLD 3 paper, I think machine learning and other ‘artificial intelligence’ approaches could also help us identify ambiguous features from frustrating fossil fragments.